- Record: found
- Abstract: found
- Article: found
RNA kinase CLP1/Cbc regulates meiosis initiation in spermatogenesis
Read this article at
Abstract
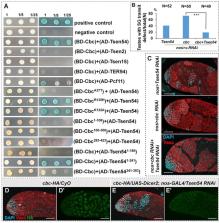
CLP1, TSEN complex, and VCP are evolutionarily conserved proteins whose mutations are associated with neurodegenerative diseases. In this study, we have found that they are also involved in germline differentiation. To optimize both quantity and quality in gametes production, germ cells expand themselves through limited mitotic cycles prior to meiosis. Stemming from our previous findings on the correlation between mRNA 3′-processing and meiosis entry, here we identify that the RNA kinase Cbc, the Drosophila member of the highly conserved CLP1 family, is a component of the program regulating the transition from mitosis to meiosis. Using genetic manipulations in Drosophila testis, we demonstrate that nuclear Cbc is required to promote meiosis entry. Combining biochemical and genetic methods, we reveal that Cbc physically and/or genetically intersects with Tsen54 and TER94 (VCP ortholog) in this process. The C-terminal half of Tsen54 is both necessary and sufficient for its binding with Cbc. Further, we illustrate the functional conservation between Cbc and mammalian CLP1 in the assays of subcellular localization and Drosophila fertility. As CLP1, TSEN complex, and VCP have also been identified in neurodegenerations of animal models, a mechanism involving these factors seems to be shared in gametogenesis and neurogenesis.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
CDD/SPARCLE: the conserved domain database in 2020
- Record: found
- Abstract: found
- Article: not found
An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases.
- Record: found
- Abstract: found
- Article: not found