- Record: found
- Abstract: found
- Article: found
The effect of environmental calcium on gene expression, biofilm formation and virulence of Vibrio parahaemolyticus
Read this article at
Abstract
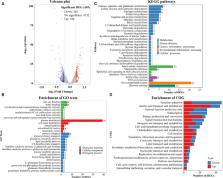
Calcium (Ca 2+) can regulate the swarming motility and virulence of Vibrio parahaemolyticus BB22. However, the effects of Ca 2+ on the physiology of V. parahaemolyticus RIMD2210633, whose genomic composition is quite different with that of BB22, have not been investigated. In this study, the results of phenotypic assays showed that the biofilm formation, c-di-GMP production, swimming motility, zebrafish survival rate, cytoxicity against HeLa cells, and adherence activity to HeLa cells of V. parahaemolyticus RIMD2210633 were significantly enhanced by Ca 2+. However, Ca 2+ had no effect on the growth, swarming motility, capsular polysaccharide (CPS) phase variation and hemolytic activity. The RNA sequencing (RNA-seq) assay disclosed 459 significantly differentially expressed genes (DEGs) in response to Ca 2+, including biofilm formation-associated genes and those encode virulence factors and putative regulators. DEGs involved in polar flagellum and T3SS1 were upregulated, whereas majority of those involved in regulatory functions and c-di-GMP metabolism were downregulated. The work helps us understand how Ca 2+ affects the behavior and gene expression of V. parahaemolyticus RIMD2210633.
Related collections
Most cited references65

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
KEGG: kyoto encyclopedia of genes and genomes.

- Record: found
- Abstract: found
- Article: found