- Record: found
- Abstract: found
- Article: found
A Novel NAC Transcription Factor From Eucalyptus, EgNAC141, Positively Regulates Lignin Biosynthesis and Increases Lignin Deposition
Read this article at
Abstract
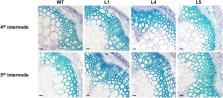
Wood formation is a complicated process under the control of a large set of transcription factors. NAC transcription factors are considered “master switches” in this process. However, few NAC members have been cloned and characterized in Eucalyptus, which is one of the most economically important woody plants. Here, we reported an NAC transcription factor from Eucalyptus grandis, EgNAC141, which has no Arabidopsis orthologs associated with xylogenesis-related processes. EgNAC141 was predominantly expressed in lignin-rich tissues, such as the stem and xylem. Overexpression of EgNAC141 in Arabidopsis resulted in stronger lignification, larger xylem, and higher lignin content. The expression of lignin biosynthetic genes in transgenic plants was significantly higher compared with wild-type plants. The transient expression of EgNAC141 activated the expression of Arabidopsis lignin biosynthetic genes in a dual-luciferase assay. Overall, these results showed that EgNAC141 is a positive regulator of lignin biosynthesis and may help us understand the regulatory mechanism of wood formation.
Related collections
Most cited references40

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
- Record: found
- Abstract: found
- Article: not found