- Record: found
- Abstract: found
- Article: found
Analysis of an IncR Plasmid Carrying bla NDM-1 Linked to an Azithromycin Resistance Region in Enterobacter hormaechei Involved in an Outbreak in Quebec
Read this article at
ABSTRACT
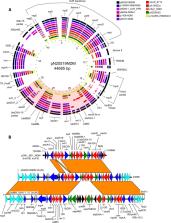
In the context of a recent rise in prevalence of NDM-encoding carbapenemase-producing Enterobacterales (CPE) in the province of QC, Canada, the genetic environment of bla NDM-1 was investigated. Three NDM-producing clinical isolates of Enterobacter hormaechei recovered from hospitalized patients involved in a putative outbreak were further characterized by whole-genome sequencing (WGS). Two isolates were confirmed by pulsed-field gel electrophoresis and WGS to be closely related. In addition to a ∼128 kb IncFII conjugative multidrug-resistance (MDR) plasmid, these isolates possessed a ∼45 kb mobilizable IncR MDR plasmid containing 2 MDR regions: a complex class 1 integron harboring bla NDM-1 and 7 other AMR genes, and the IS26-mph(A) -mrx-mphR(A) -IS6100 azithromycin resistance unit. The predicted antimicrobial resistance (AMR) genes correlated with the antimicrobial susceptibility testing results. The multidrug-resistant phenotype in addition to the presence of two important mobile genetic elements, suggest a potent role as a reservoir of antibiotic resistance for such a small IncR plasmid.
IMPORTANCE Analyzing the genetic environment of clinically relevant MDR genes can provide information on the way in which such genes are maintained and disseminated. Understanding this phenomenon is of interest for clinicians as it can also provide insight on where these genes might have been sourced, possibly supporting outbreak investigations.
Related collections
Most cited references12

- Record: found
- Abstract: found
- Article: found
Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads
- Record: found
- Abstract: found
- Article: not found
Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae

- Record: found
- Abstract: found
- Article: found