- Record: found
- Abstract: found
- Article: found
A longitudinal field study of commercial honey bees shows that non-native probiotics do not rescue antibiotic treatment, and are generally not beneficial
Read this article at
Abstract
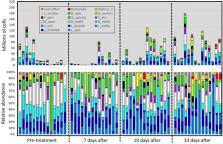
Probiotics are widely used in agriculture including commercial beekeeping, but there is little evidence supporting their effectiveness. Antibiotic treatments can greatly distort the gut microbiome, reducing its protective abilities and facilitating the growth of antibiotic resistant pathogens. Commercial beekeepers regularly apply antibiotics to combat bacterial infections, often followed by an application of non-native probiotics advertised to ease the impact of antibiotic-induced gut dysbiosis. We tested whether probiotics affect the gut microbiome or disease prevalence, or rescue the negative effects of antibiotic induced gut dysbiosis. We found no difference in the gut microbiome or disease markers by probiotic application or antibiotic recovery associated with probiotic treatment. A colony-level application of the antibiotics oxytetracycline and tylosin produced an immediate decrease in gut microbiome size, and over the longer-term, very different and persistent dysbiotic effects on the composition and membership of the hindgut microbiome. Our results demonstrate the lack of probiotic effect or antibiotic rescue, detail the duration and character of dysbiotic states resulting from different antibiotics, and highlight the importance of the gut microbiome for honeybee health.
Related collections
Most cited references79
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities.

- Record: found
- Abstract: found
- Article: found