- Record: found
- Abstract: found
- Article: found
Long noncoding RNA MALAT1 inhibits the apoptosis and autophagy of hepatocellular carcinoma cell by targeting the microRNA-146a/PI3K/Akt/mTOR axis
Read this article at
Abstract
Background
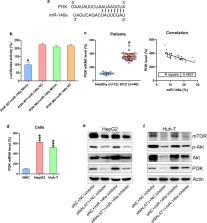
Increased long noncoding RNA (lncRNA) expression is characteristic to hepatocellular carcinoma (HCC) and several other neoplasms. The present study aimed to identify the mechanism underlying modulation of HCC development by the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1).
Methods
Quantitative real-time polymerase chain reaction was used to determine MALAT1 and microRNA (miR)-146a expression in HCC tissues and cell lines. Western blotting was performed to measure PI3K, Akt, and mTOR levels. Dual-luciferase reporter assay was used to validate the direct targeting and negative regulatory interaction between miR-146a and MALAT1. Cell viability, proliferation, and apoptosis were analyzed using an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay, colony formation assay, and flow cytometry, respectively; autophagy was detected based on LC3B expression.
Results
MALAT1 expression was higher in HCC tissues than in normal tissues. MALAT1 upregulation promoted HCC cell proliferation, whereas MALAT1 downregulation promoted HCC apoptosis and autophagy. Moreover, effects of MALAT1 downregulation on HCC cells were abolished by miR-146a inhibition. miR-146a directly targeted the 3′-untranslated region of PI3K, and PI3K protein level was clearly decreased upon miR-146a mimic transfection.
Related collections
Most cited references33
- Record: found
- Abstract: not found
- Article: not found
Management of hepatocellular carcinoma.
- Record: found
- Abstract: found
- Article: not found
Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer.
- Record: found
- Abstract: found
- Article: not found