- Record: found
- Abstract: found
- Article: found
Analysis of the translatome in solid tumors using polyribosome profiling/RNA-Seq
Read this article at
Abstract
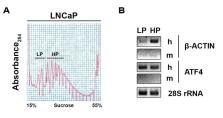
Gene expression involves multiple steps from the transcription of a mRNA in the nucleus to the production of the encoded protein in the cytoplasm. This final step occurs through a highly regulated process of mRNA translation on ribosomes that is required to maintain cell homeostasis. Alterations in the control of mRNA translation may lead to cell’s transformation, a hallmark of cancer development. Indeed, recent advances indicated that increased translation of mRNAs encoding tumor-promoting proteins may be a key mechanism of tumor resistance in several cancers. Moreover, it was found that proteins whose encoding mRNAs are translated at higher efficiencies may be effective biomarkers. Evaluation of global changes in translation efficiency in human tumors has thus the potential of better understanding what can be used as biomarkers and therapeutic targets. Investigating changes in translation efficiency in human cancer cells has been made possible through the development and use of the polyribosome profiling combined with DNA microarray or deep RNA sequencing (RNA-Seq). While helpful, the use of cancer cell lines has many limitations and it is essential to define translational changes in human tumor samples in order to properly prioritize genes implicated in cancer phenotype. We present an optimized polyribosome RNA-Seq protocol suitable for quantitative analysis of mRNA translation that occurs in human tumor samples and murine xenografts. Applying this innovative approach to human tumors, which requires a complementary bioinformatics analysis, unlocks the potential to identify key mRNA which are preferentially translated in tumor tissue compared to benign tissue as well as translational changes which occur following treatment. These technical advances will be of interest to those researching all solid tumors, opening possibilities for understanding what may be therapeutic Achilles heels’ or relevant biomarkers.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Translational control by 5'-untranslated regions of eukaryotic mRNAs.
- Record: found
- Abstract: found
- Article: not found