- Record: found
- Abstract: found
- Article: found
Molecular Docking: Shifting Paradigms in Drug Discovery.
Read this article at
Abstract
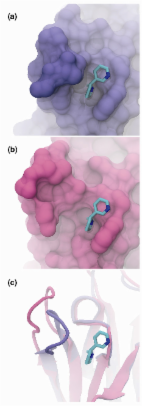
Molecular docking is an established in silico structure-based method widely used in drug discovery. Docking enables the identification of novel compounds of therapeutic interest, predicting ligand-target interactions at a molecular level, or delineating structure-activity relationships (SAR), without knowing a priori the chemical structure of other target modulators. Although it was originally developed to help understanding the mechanisms of molecular recognition between small and large molecules, uses and applications of docking in drug discovery have heavily changed over the last years. In this review, we describe how molecular docking was firstly applied to assist in drug discovery tasks. Then, we illustrate newer and emergent uses and applications of docking, including prediction of adverse effects, polypharmacology, drug repurposing, and target fishing and profiling, discussing also future applications and further potential of this technique when combined with emergent techniques, such as artificial intelligence.
Related collections
Most cited references156
- Record: found
- Abstract: found
- Article: not found
A fast flexible docking method using an incremental construction algorithm.
- Record: found
- Abstract: found
- Article: found