- Record: found
- Abstract: found
- Article: found
MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences
Read this article at
Abstract
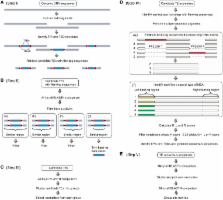
Miniature inverted-repeat transposable elements (MITEs) are a special type of Class 2 non-autonomous transposable element (TE) that are abundant in the non-coding regions of the genes of many plant and animal species. The accurate identification of MITEs has been a challenge for existing programs because they lack coding sequences and, as such, evolve very rapidly. Because of their importance to gene and genome evolution, we developed MITE-Hunter, a program pipeline that can identify MITEs as well as other small Class 2 non-autonomous TEs from genomic DNA data sets. The output of MITE-Hunter is composed of consensus TE sequences grouped into families that can be used as a library file for homology-based TE detection programs such as RepeatMasker. MITE-Hunter was evaluated by searching the rice genomic database and comparing the output with known rice TEs. It discovered most of the previously reported rice MITEs (97.6%), and found sixteen new elements. MITE-Hunter was also compared with two other MITE discovery programs, FINDMITE and MUST. Unlike MITE-Hunter, neither of these programs can search large genomic data sets including whole genome sequences. More importantly, MITE-Hunter is significantly more accurate than either FINDMITE or MUST as the vast majority of their outputs are false-positives.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
The Sorghum bicolor genome and the diversification of grasses.
- Record: found
- Abstract: found
- Article: not found
An active DNA transposon family in rice.
- Record: found
- Abstract: found
- Article: not found