- Record: found
- Abstract: found
- Article: found
RosBREED: bridging the chasm between discovery and application to enable DNA-informed breeding in rosaceous crops
Read this article at
Abstract
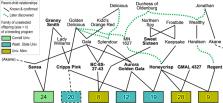
The Rosaceae crop family (including almond, apple, apricot, blackberry, peach, pear, plum, raspberry, rose, strawberry, sweet cherry, and sour cherry) provides vital contributions to human well-being and is economically significant across the U.S. In 2003, industry stakeholder initiatives prioritized the utilization of genomics, genetics, and breeding to develop new cultivars exhibiting both disease resistance and superior horticultural quality. However, rosaceous crop breeders lacked certain knowledge and tools to fully implement DNA-informed breeding—a “chasm” existed between existing genomics and genetic information and the application of this knowledge in breeding. The RosBREED project (“Ros” signifying a Rosaceae genomics, genetics, and breeding community initiative, and “BREED”, indicating the core focus on breeding programs), addressed this challenge through a comprehensive and coordinated 10-year effort funded by the USDA-NIFA Specialty Crop Research Initiative. RosBREED was designed to enable the routine application of modern genomics and genetics technologies in U.S. rosaceous crop breeding programs, thereby enhancing their efficiency and effectiveness in delivering cultivars with producer-required disease resistances and market-essential horticultural quality. This review presents a synopsis of the approach, deliverables, and impacts of RosBREED, highlighting synergistic global collaborations and future needs. Enabling technologies and tools developed are described, including genome-wide scanning platforms and DNA diagnostic tests. Examples of DNA-informed breeding use by project participants are presented for all breeding stages, including pre-breeding for disease resistance, parental and seedling selection, and elite selection advancement. The chasm is now bridged, accelerating rosaceous crop genetic improvement.
Related collections
Most cited references193
- Record: found
- Abstract: found
- Article: not found
Efficient methods to compute genomic predictions.

- Record: found
- Abstract: found
- Article: found
JBrowse: a dynamic web platform for genome visualization and analysis
- Record: found
- Abstract: found
- Article: not found