- Record: found
- Abstract: found
- Article: found
System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection

Read this article at
Summary
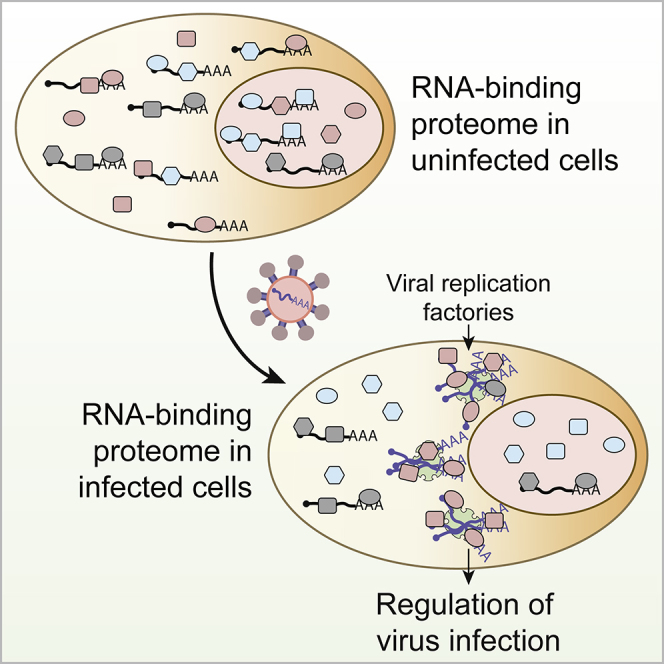
The compendium of RNA-binding proteins (RBPs) has been greatly expanded by the development of RNA-interactome capture (RIC). However, it remained unknown if the complement of RBPs changes in response to environmental perturbations and whether these rearrangements are important. To answer these questions, we developed “comparative RIC” and applied it to cells challenged with an RNA virus called sindbis (SINV). Over 200 RBPs display differential interaction with RNA upon SINV infection. These alterations are mainly driven by the loss of cellular mRNAs and the emergence of viral RNA. RBPs stimulated by the infection redistribute to viral replication factories and regulate the capacity of the virus to infect. For example, ablation of XRN1 causes cells to be refractory to SINV, while GEMIN5 moonlights as a regulator of SINV gene expression. In summary, RNA availability controls RBP localization and function in SINV-infected cells.
Graphical Abstract
Highlights
Abstract
Garcia-Moreno, Noerenberg, Ni, and colleagues developed “comparative RNA-interactome capture” to analyze the RNA-bound proteome during virus infection. More than 200 cellular RNA-binding proteins change their binding activity in response to this challenge, mainly driven by transcript availability. Many of these RNA-binding proteins regulate viral replication and can be targeted to influence infection outcome.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA.
- Record: found
- Abstract: found
- Article: not found
The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity
- Record: found
- Abstract: found
- Article: not found