- Record: found
- Abstract: found
- Article: found
The Major Risk Factors for Alzheimer’s Disease: Age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques

SUMMARY
Gene expression profiles of more than 10,000 individual microglial cells isolated from cortex and hippocampus of male and female App NL-G-F mice over time demonstrate that progressive amyloid-b accumulation accelerates two main activated microglia states that are also present during normal aging. Activated response microglia (ARMs) are composed of specialized subgroups overexpressing MHC type II and putative tissue repair genes ( Dkk2, Gpnmb, and Spp1) and are strongly enriched with Alzheimer’s disease (AD) risk genes. Microglia from female mice progress faster in this activation trajectory. Similar activated states are also found in a second AD model and in human brain. Apoe, the major genetic risk factor for AD, regulates the ARMs but not the interferon response microglia (IRMs). Thus, the ARMs response is the converging point for aging, sex, and genetic AD risk factors.
Graphical Abstract
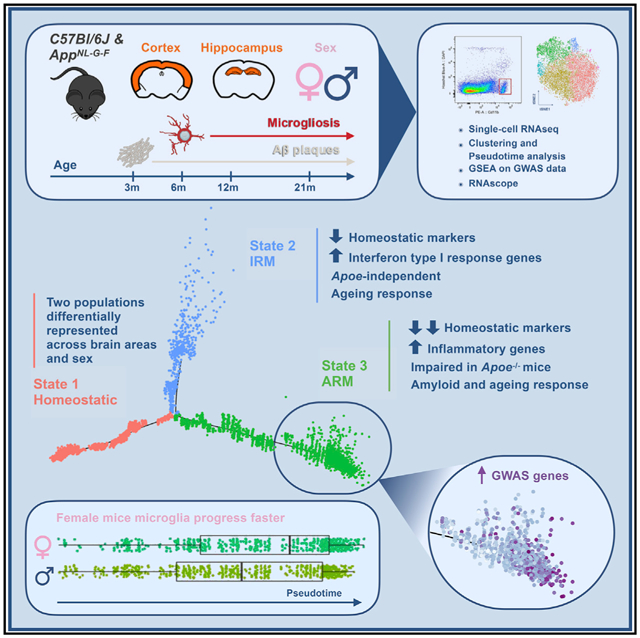
In Brief
Sala Frigerio et al. show how microglia respond to amyloid-b, the Alzheimer’s disease (AD)-causing factor. Their major response, the ARMs response, is enriched for AD risk genes, is abolished by Apoe deletion, develops faster in female mice, and is also part of normal aging. Thus, major AD risk factors converge on microglia.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families.
- Record: found
- Abstract: found
- Article: not found
TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia.
- Record: found
- Abstract: found
- Article: not found