- Record: found
- Abstract: found
- Article: found
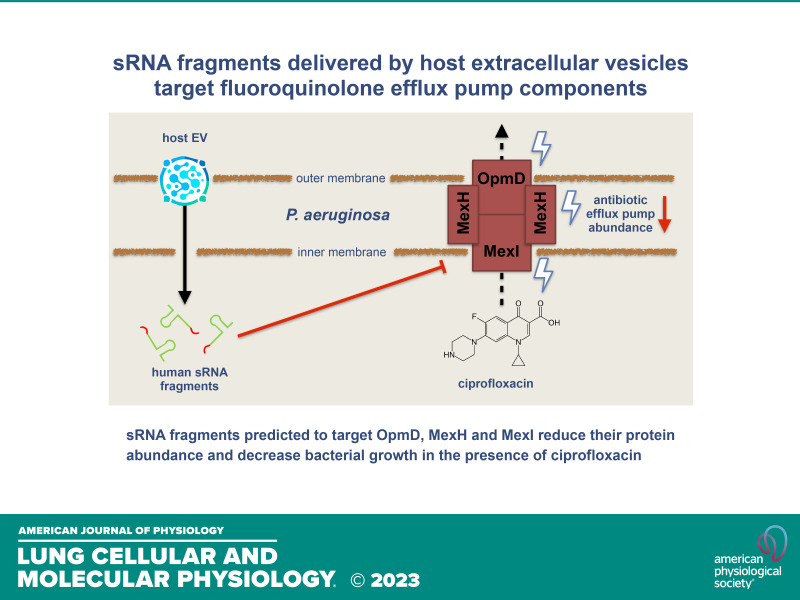
An rRNA fragment in extracellular vesicles secreted by human airway epithelial cells increases the fluoroquinolone sensitivity of P. aeruginosa

Read this article at
Abstract
Abstract
Lung infections caused by antibiotic-resistant strains of Pseudomonas aeruginosa are difficult to eradicate in immunocompromised hosts such as those with cystic fibrosis. We previously demonstrated that extracellular vesicles (EVs) secreted by primary human airway epithelial cells (AECs) deliver microRNA let-7b-5p to P. aeruginosa to suppress biofilm formation and increase sensitivity to beta-lactam antibiotics. In this study, we show that EVs secreted by AECs transfer multiple distinct short RNA fragments to P. aeruginosa that are predicted to target the three subunits of the fluoroquinolone efflux pump MexHI-OpmD, thus increasing antibiotic sensitivity. Exposure of P. aeruginosa to EVs resulted in a significant reduction in the protein levels of MexH (−48%), MexI (−50%), and OpmD (−35%). Moreover, EVs reduced planktonic growth of P. aeruginosa in the presence of the fluoroquinolone antibiotic ciprofloxacin by 20%. A mexGHI-opmD deletion mutant of P. aeruginosa phenocopied this increased sensitivity to ciprofloxacin. Finally, we found that a fragment of an 18S ribosomal RNA (rRNA) external transcribed spacer that was transferred to P. aeruginosa by EVs reduced planktonic growth of P. aeruginosa in the presence of ciprofloxacin, reduced the minimum inhibitory concentration of P. aeruginosa for ciprofloxacin by over 50%, and significantly reduced protein levels of both MexH and OpmD. In conclusion, an rRNA fragment secreted by AECs in EVs that targets the fluoroquinolone efflux pump MexHI-OpmD downregulated these proteins and increased the ciprofloxacin sensitivity of P. aeruginosa. A combination of rRNA fragments and ciprofloxacin packaged in nanoparticles or EVs may benefit patients with ciprofloxacin-resistant P. aeruginosa infections.
NEW & NOTEWORTHY Human RNA fragments transported in extracellular vesicles interfere with Pseudomonas aeruginosa drug efflux pumps. A combination of rRNA fragments and ciprofloxacin packaged in nanoparticles or EVs may benefit patients with antibiotic-resistant P. aeruginosa infections.
Related collections
Most cited references85

- Record: found
- Abstract: found
- Article: found
fastp: an ultra-fast all-in-one FASTQ preprocessor

- Record: found
- Abstract: found
- Article: found
BLAST+: architecture and applications

- Record: found
- Abstract: found
- Article: found