- Record: found
- Abstract: found
- Article: found
Whole exome sequencing establishes diagnosis of Charcot–Marie–Tooth 4J, 1C, and X1 subtypes

Read this article at
Abstract
Background
Charcot–Marie–Tooth (CMT) hereditary polyneuropathies pose a diagnostic challenge. Our aim here is to describe CMT patients diagnosed by whole exome sequencing (WES) following years of fruitless testing.
Methods/Results
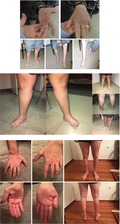
Three patients with polyneuropathy suspected to be genetic in origin, but not harboring PMP22 gene deletion/duplication, were offered WES. The first patient, a 66‐year‐old man, had been suffering from progressive weakness and atrophies in the lower and upper extremities for 20 years. Due to ambiguous electrophysiological findings, immune therapies were administered to no avail. Twelve years after PMP22 deletion/duplication testing, WES revealed two pathogenic variants in the FIG4 gene (p.Ile41Thr and p.Phe598fs, respectively), as a cause of CMT 4J. The second patient, a 19‐year‐old man, had been suffering from hearing and gait impairment since at least his infancy, and recently presented with weakness and dystonia of the lower extremities. In this patient, WES identified the p.Leu122Val LITAF gene variant in heterozygous state, suggesting the diagnosis of CMT 1C, several years after initial genetic analyses. The third patient, a 44‐year‐old man, presented with progressive weakness and atrophies of the lower and upper extremities since the age of 17 years old. In this patient, WES identified the hemizygous p.Arg164Gln pathogenic variant in the GJB1 gene, establishing the diagnosis of CMT X1, 8 years after testing for PMP22 deletion/duplication.
Abstract
Here we describe three cases of Charcot–Marie–Tooth hereditary polyneuropathy (subtypes 4J, 1C, and X1, respectively) that we have diagnosed using whole exome sequencing (WES) after several years of nondiagnostic genetic and nongenetic tests. These cases demonstrate the advantages of a simplified strategy in diagnosing hereditary peripheral neuropathies, namely proceeding directly from PMP22 deletion/duplication analysis to WES for the diagnostic evaluation of these patients.
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: not found
Connexin mutations in X-linked Charcot-Marie-Tooth disease.

- Record: found
- Abstract: found
- Article: found
The allelic spectrum of Charcot–Marie–Tooth disease in over 17,000 individuals with neuropathy
- Record: found
- Abstract: found
- Article: not found