- Record: found
- Abstract: found
- Article: found
Spindle and Kinetochore-associated Family Genes are Prognostic and Predictive Biomarkers in Hepatocellular Carcinoma

Read this article at
Abstract
Background and Aims
Hepatocellular carcinoma (HCC) is one of the most frequent malignant tumors. Spindle and kinetochore-associated (SKA) family genes are essential for the maintenance of the metaphase plate and spindle checkpoint silencing during mitosis. Recent studies have indicated that dysregulation of SKA family genes induces tumorigenesis, tumor progression, and chemoresistance via modulation of cell cycle and DNA replication. However, the differential transcription of SKAs in the context of HCC and its prognostic significance has not been demonstrated.
Methods
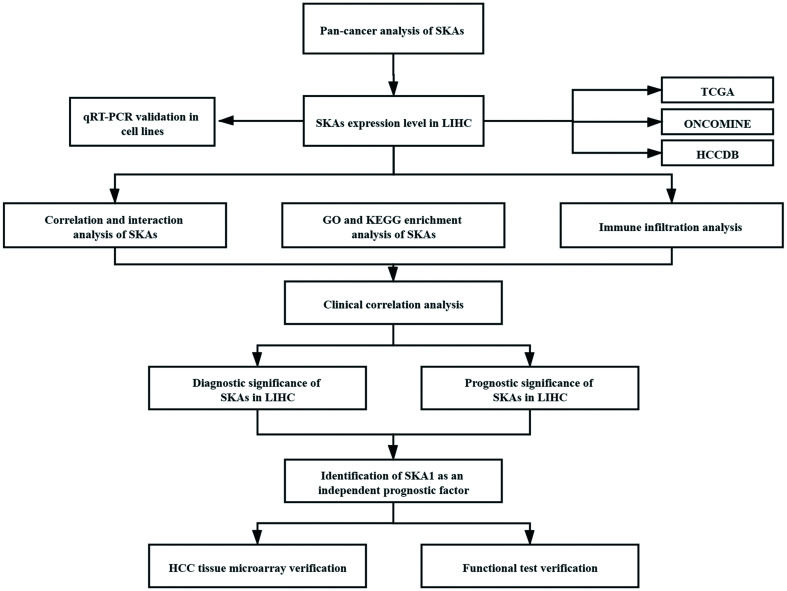
Bioinformatics analyses were performed using TCGA, ONCOMINE, HCCDB, Kaplan-Meier plotter, STRING, GEPIA databases. qRT-PCR, western blot, and functional assays were utilized for in vitro experiments.
Results
We found remarkable upregulation of transcripts of SKA family genes in HCC samples compared with normal liver samples on bioinformatics analyses and in vitro validation. Interaction analysis and enrichment analysis showed that SKA family members were mainly related to microtubule motor activity, mitosis, and cell cycle. Immuno-infiltration analysis showed a correlation of all SKA family genes with various immune cell subsets, especially T helper 2 (Th2) cells. Transcriptional levels of SKA family members were positively associated with histologic grade, T stage, and α-fetoprotein in HCC patients. Receiver operating characteristic curve analysis demonstrated a strong predictive ability of SKA1/2/3 for HCC. Increased expression of these SKAs was associated with unfavorable overall survival, progression-free survival, and disease-specific survival. On Cox proportional hazards regression analyses, SKA1 upregulation and pathological staging were independent predictors of overall survival and disease-specific survival of HCC patients. Finally, clinical tissue microarray validation and in vitro functional assays revealed SKA1 acts an important regulatory role in tumor malignant behavior.
Graphical abstract
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found