- Record: found
- Abstract: found
- Article: found
1453. PBP2, PBP2a and PBP4 Clone-specific Polymorphisms are not Associated to Ceftaroline (CPT) Susceptibility in Chilean Clinical Isolates of Methicillin-Resistant Staphylococcus aureus (MRSA)

Read this article at
Abstract
Background
CPT is a last-generation cephalosporin active against MRSA due to its affinity for PBP2a. CPT-resistance (CPT-R) is well-described, with mutations in the active transpeptidase domain of PBP2a associated to high-level resistance. The accumulation of changes in the non-penicillin-binding domain of PBP2a has been linked to elevations of the minimal inhibitory concentration (MIC) to CPT to levels around 2-4ug/mL. PBP4 and PBP2 have also been implicated as potentially relevant mecA-independent mechanisms of CPT-R. We recently reported high rates of CPT-non-susceptibility in clinical MRSA strains from Chile. However, the mutational landscape of PBPs in clinical MRSA isolates from Chile and its relation to CPT susceptibility has not been assessed.
Methods
We analyzed 180 MRSA isolates collected from 2000-2018 in Santiago, Chile. Identification was confirmed by MALDI-TOF and methicillin resistance with cefoxitin disk-diffusion. CPT susceptibility was performed by BMD following CLSI-2019 guidance. Whole-genome sequencing was performed for all isolates; the mutational profile of PBPs was determined using reference sequences for PBP2 (AGY89563.1), PBP2a (NG_047938.1) and PBP4 (X91786.1).
Results
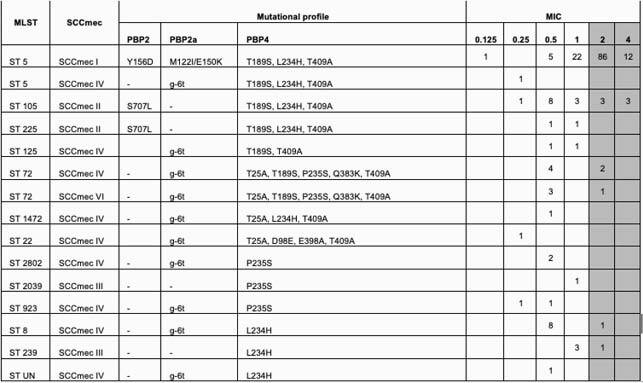
All isolates were phenotypically-confirmed MRSA and harbored mecA. The MIC 50/MIC 90 by BMD was 2/2μg/dL; only 71 (39%) isolates were CPT-susceptible (MIC <1µg/mL). Most isolates belonged to ST5/SCC mecI (70%,126/180), ST105/SCCmecII (10%,18/180) and ST8/SCC mecIV (5%, 9/180). All ST5/SCC mecI isolates carried the mutations in PBP2 (Y156D), PBP2a (M122I and E150K), and PBP4 (T189S, L234H, and T409A); CPT-susceptibility among ST5/SCC mecI was only 22%. On the other hand, all ST105/SCC mecII isolates had mutations in PBP2 (S707L) and PBP4 (T189S, L234H, and T409A) and exhibited a higher CPT-susceptibility rate (67%). All 9 isolates belonging to the ST8/SCC mecIV lineage harbored a non-coding mutation in PBP2a (g-6t) and the previously observed L234H change in PBP4. Importantly, no association between specific polymorphisms and MIC to CPT was found.
Table 1. PBPs mutations compared to CPT MICs by MLST and SCCmec