- Record: found
- Abstract: found
- Article: found
The oralome and its dysbiosis: New insights into oral microbiome-host interactions

Read this article at
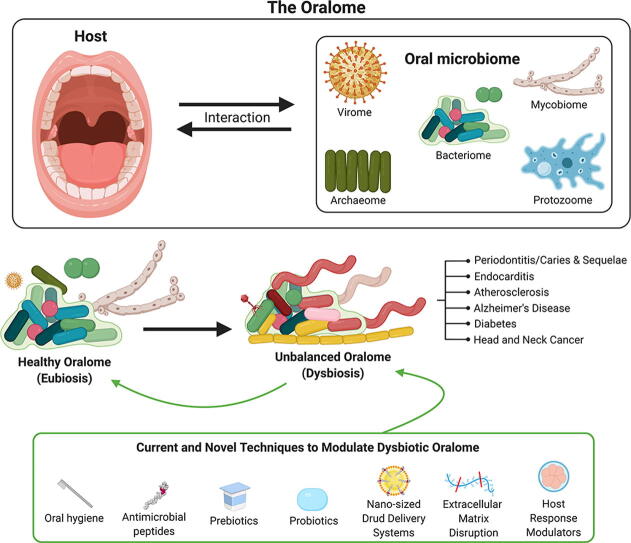
Graphical abstract
Abstract
The oralome is the summary of the dynamic interactions orchestrated between the ecological community of oral microorganisms (comprised of up to approximately 1000 species of bacteria, fungi, viruses, archaea and protozoa - the oral microbiome) that live in the oral cavity and the host. These microorganisms form a complex ecosystem that thrive in the dynamic oral environment in a symbiotic relationship with the human host. However, the microbial composition is significantly affected by interspecies and host-microbial interactions, which in turn, can impact the health and disease status of the host. In this review, we discuss the composition of the oralome and inter-species and host-microbial interactions that take place in the oral cavity and examine how these interactions change from healthy (eubiotic) to disease (dysbiotic) states. We further discuss the dysbiotic signatures associated with periodontitis and caries and their sequalae, (e.g., tooth/bone loss and pulpitis), and the systemic diseases associated with these oral diseases, such as infective endocarditis, atherosclerosis, diabetes, Alzheimer’s disease and head and neck/oral cancer. We then discuss current computational techniques to assess dysbiotic oral microbiome changes. Lastly, we discuss current and novel techniques for modulation of the dysbiotic oral microbiome that may help in disease prevention and treatment, including standard hygiene methods, prebiotics, probiotics, use of nano-sized drug delivery systems (nano-DDS), extracellular polymeric matrix (EPM) disruption, and host response modulators.
Related collections
Most cited references367

- Record: found
- Abstract: found
- Article: found
Revised Estimates for the Number of Human and Bacteria Cells in the Body
- Record: found
- Abstract: found
- Article: not found
The bactericidal effect of silver nanoparticles.
- Record: found
- Abstract: found
- Article: not found