- Record: found
- Abstract: found
- Article: found
Possible mechanisms of lycopene amelioration of learning and memory impairment in rats with vascular dementia

Read this article at
Abstract
Abstract
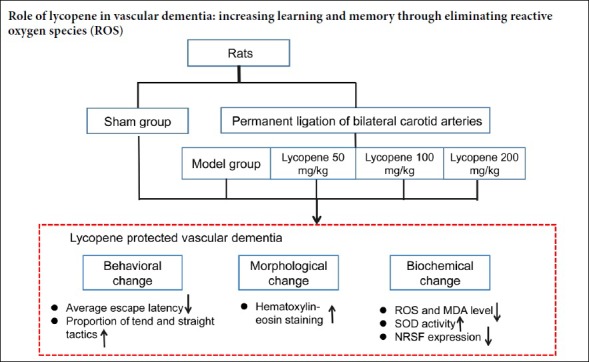
Oxidative stress is involved in the pathogenesis of vascular dementia. Studies have shown that lycopene can significantly inhibit oxidative stress; therefore, we hypothesized that lycopene can reduce the level of oxidative stress in vascular dementia. A vascular dementia model was established by permanent bilateral ligation of common carotid arteries. The dosage groups were treated with lycopene (50, 100 and 200 mg/kg) every other day for 2 months. Rats without bilateral carotid artery ligation were prepared as a sham group. To test the ability of learning and memory, the Morris water maze was used to detect the average escape latency and the change of search strategy. Hematoxylin-eosin staining was used to observe changes of hippocampal neurons. The levels of oxidative stress factors, superoxide dismutase and malondialdehyde, were measured in the hippocampus by biochemical detection. The levels of reactive oxygen species in the hippocampus were observed by dihydroethidium staining. The distribution and expression of oxidative stress related protein, neuron-restrictive silencer factor, in hippocampal neurons were detected by immunofluorescence histochemistry and western blot assays. After 2 months of drug administration, (1) in the model group, the average escape latency was longer than that of the sham group, and the proportion of straight and tend tactics was lower than that of the sham group, and the hippocampal neurons were irregularly arranged and the cytoplasm was hyperchromatic. (2) The levels of reactive oxygen species and malondialdehyde in the hippocampus of the model group rats were increased, and the activity of superoxide dismutase was decreased. (3) Lycopene (50, 100 and 200 mg/kg) intervention improved the above changes, and the lycopene 100 mg/kg group showed the most significant improvement effect. (4) Neuron-restrictive silencer factor expression in the hippocampus was lower in the sham group and the lycopene 100 mg/kg group than in the model group. (5) The above data indicate that lycopene 100 mg/kg could protect against the learning-memory ability impairment of vascular dementia rats. The protective mechanism was achieved by inhibiting oxidative stress in the hippocampus. The experiment was approved by the Animal Ethics Committee of Fujian Medical University, China (approval No. 2014-025) in June 2014.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons.
- Record: found
- Abstract: found
- Article: not found
Role of brain-derived neurotrophic factor in Huntington's disease.
- Record: found
- Abstract: found
- Article: not found